The nucleosome tethering kit enables the tethering of nucleosomal arrays in optical tweezers to study the interactions and regulation between DNA binding proteins and nucleosomes.
The kit includes biotinylated 6.3 kb DNA handles (exact size 6,298 bp) and the pUC-LUMICKS-601 plasmid. The plasmid contains a 12-repeat array of a 172 bp clone-601 DNA sequence flanked by restriction sites for the type IIS restriction enzyme BsaI. Upon digestion with BsaI, the DNA repeat array can be purified from the plasmid, loaded with histones, and finally ligated to the 6.3 kb handles.
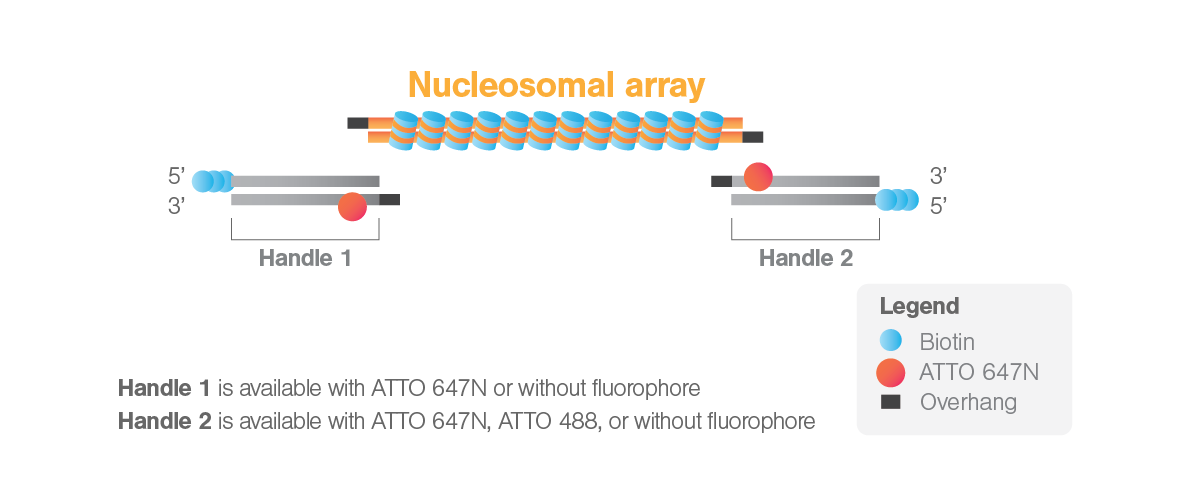
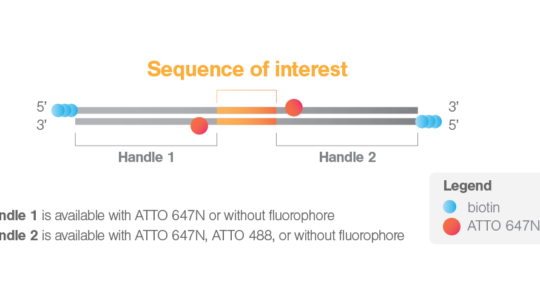
The ligation is designed so that DNA handle 1 will be ligated at the 5′ end of the nucleosomal array, and DNA handle 2 will be ligated at the 3′ end. Additionally, the kit offers different versions of handles with or without ATTO fluorophores. The options for handle 1 include with ATTO 647N or without a fluorophore, while for handle 2, the options are with ATTO 647N, ATTO 488, or without a fluorophore. The combination of two handles without fluorophores is offered here for a discounted early release price on this page.
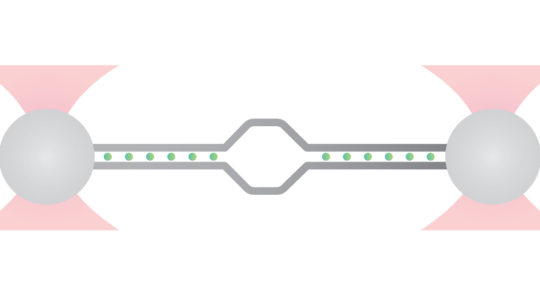
Once the nucleosomes are ligated to the two DNA handles, the biotin moieties of each DNA handle will allow tethering of the nucleosomes between two streptavidin-coated beads trapped in optical tweezers (Figure 1). In this configuration, if handles with fluorophores are chosen, the fluorophores will flank the nucleosomes (11 bps from the ligation sites) and can be used to position the SOI on the focal plane before incubation with fluorescent proteins. This enables the setup of optimal fluorescence imaging conditions before starting a DNA-protein interaction measurement, which allows capturing even the first interactions. By choosing handles labeled differently, an asymmetric configuration with the nucleosomal array can be achieved, and the 5′-3′ directionality can be determined when tethered in the C-Trap. Additionally, in case both handles are labeled with ATTO fluorophores, the known distance between the two fluorophores can be used as a ruler on the tethered DNA to precisely determine the position on the DNA sequence of fluorescent proteins interacting with the sequence of interest.
In addition to the DNA handles and pUC-LUMICKS_601 plasmid, the kit includes a 3 kbp control DNA with overhangs (exact size 3,065 bp). As nucleosome preparations are typically performed in a buffer containing a relatively high concentration of salt (>0.1 M NaCl), we recommend using Salt-T4® DNA Ligase from New England BioLabs (supplied by the user; catalog #M0467) for the ligation of the nucleosomes to the DNA handles.
The kit enables 4 reactions. Each reaction can be used for at least 5 experimental sessions on the C-Trap.
Estimated 1-4 weeks delivery time.